How shut can an open mannequin get to AlphaFold3-level accuracy when it matches coaching knowledge, mannequin scale and inference funds? ByteDance has launched Protenix-v1, a complete AlphaFold3 (AF3) copy for biomolecular construction prediction, launched with code and mannequin parameters underneath Apache 2.0. The mannequin targets AF3-level efficiency throughout protein, DNA, RNA and ligand constructions whereas protecting your entire stack open and extensible for analysis and manufacturing.
The core launch additionally ships with PXMeter v1.0.0, an analysis toolkit and dataset suite for clear benchmarking on greater than 6k complexes with time-split and domain-specific subsets.
What’s Protenix-v1?
Protenix is described as ‘Protenix: Protein + X‘, a basis mannequin for high-accuracy biomolecular construction prediction. It predicts all-atom 3D constructions for complexes that may embody:
- Proteins
- Nucleic acids (DNA and RNA)
- Small-molecule ligands
The analysis workforce defines Protenix as a complete AF3 copy. It re-implements the AF3-style diffusion structure for all-atom complexes and exposes it in a trainable PyTorch codebase.
The challenge is launched as a full stack:
- Coaching and inference code
- Pre-trained mannequin weights
- Information and MSA pipelines
- A browser-based Protenix Net Server for interactive use
AF3-level efficiency underneath matched constraints
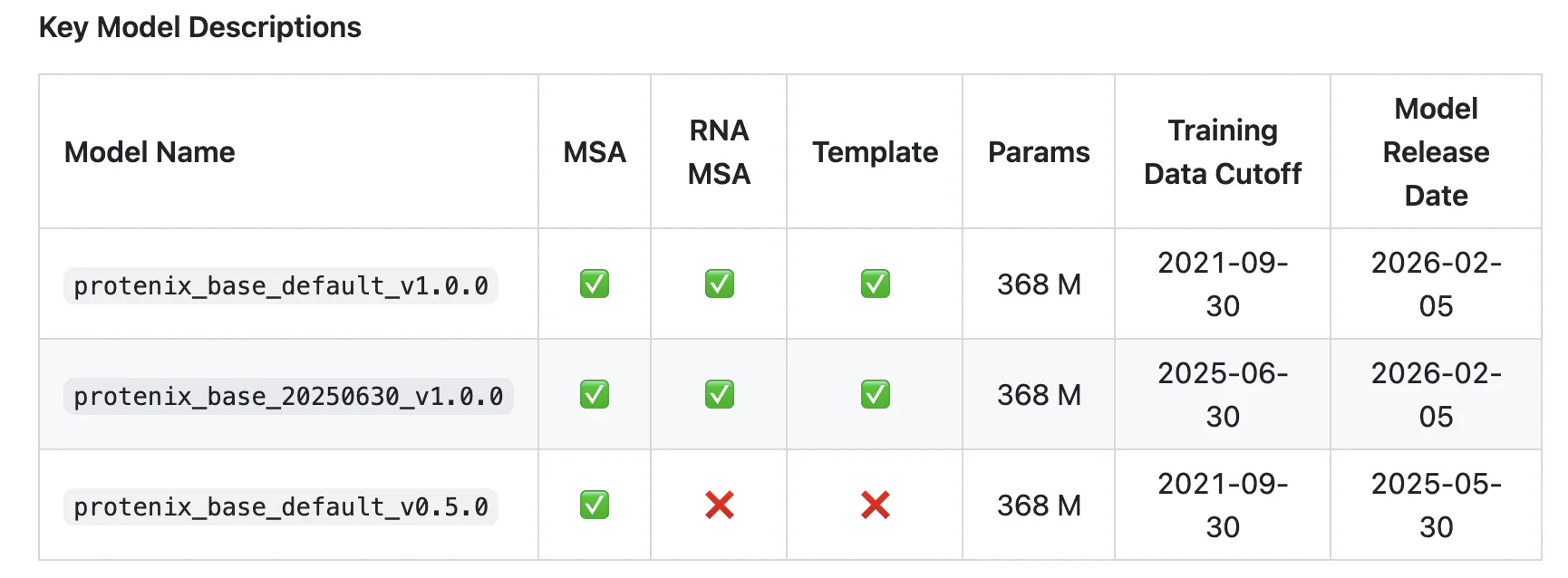
As per the analysis workforce Protenix-v1 (protenix_base_default_v1.0.0) is ‘the primary totally open-source mannequin that outperforms AlphaFold3 throughout various benchmark units whereas adhering to the identical coaching knowledge cutoff, mannequin scale, and inference funds as AlphaFold3.‘
The essential constraints are:
- Coaching knowledge cutoff: 2021-09-30, aligned with AF3’s PDB cutoff.
- Mannequin scale: Protenix-v1 itself has 368M parameters; AF3 scale is matched however not disclosed.
- Inference funds: comparisons use related sampling budgets and runtime constraints.
On difficult targets comparable to antigen–antibody complexes, rising the variety of sampled candidates from a number of to a whole lot yields constant log-linear enhancements in accuracy. This offers a transparent and documented inference-time scaling conduct relatively than a single fastened working level.
PXMeter v1.0.0: Analysis for 6k+ complexes
To assist these claims, the analysis workforce launched PXMeter v1.0.0, an open-source toolkit for reproducible construction prediction benchmarks.
PXMeter supplies:
- A manually curated benchmark dataset, with non-biological artifacts and problematic entries eliminated
- Time-split and domain-specific subsets (for instance, antibody–antigen, protein–RNA, ligand complexes)
- A unified analysis framework that computes metrics comparable to advanced LDDT and DockQ throughout fashions
The related PXMeter analysis paper, ‘Revisiting Construction Prediction Benchmarks with PXMeter,‘ evaluates Protenix, AlphaFold3, Boltz-1 and Chai-1 on the identical curated duties, and exhibits how totally different dataset designs have an effect on mannequin rating and perceived efficiency.
How Protenix matches into the broader stack?
Protenix is a part of a small ecosystem of associated tasks:
- PXDesign: a binder design suite constructed on the Protenix basis mannequin. It studies 20–73% experimental hit charges and 2–6× increased success than strategies comparable to AlphaProteo and RFdiffusion, and is accessible through the Protenix Server.
- Protenix-Dock: a classical protein–ligand docking framework that makes use of empirical scoring capabilities relatively than deep nets, tuned for inflexible docking duties.
- Protenix-Mini and follow-on work comparable to Protenix-Mini+: light-weight variants that cut back inference value utilizing architectural compression and few-step diffusion samplers, whereas protecting accuracy inside just a few p.c of the complete mannequin on commonplace benchmarks.
Collectively, these elements cowl construction prediction, docking, and design, and share interfaces and codecs, which simplifies integration into downstream pipelines.
Key Takeaways
- AF3-class, totally open mannequin: Protenix-v1 is an AF3-style all-atom biomolecular construction predictor with open code and weights underneath Apache 2.0, focusing on proteins, DNA, RNA and ligands.
- Strict AF3 alignment for truthful comparability: Protenix-v1 matches AlphaFold3 on crucial axes: coaching knowledge cutoff (2021-09-30), mannequin scale class and comparable inference funds, enabling truthful AF3-level efficiency claims.
- Clear benchmarking with PXMeter v1.0.0: PXMeter supplies a curated benchmark suite over 6k+ complexes with time-split and domain-specific subsets plus unified metrics (for instance, advanced LDDT, DockQ) for reproducible analysis.
- Verified inference-time scaling conduct: Protenix-v1 exhibits log-linear accuracy good points because the variety of sampled candidates will increase, giving a documented latency–accuracy trade-off relatively than a single fastened working level.
Take a look at the Repo and Attempt it right here. Additionally, be happy to comply with us on Twitter and don’t overlook to affix our 100k+ ML SubReddit and Subscribe to our E-newsletter. Wait! are you on telegram? now you may be part of us on telegram as nicely.


